| Home | Sources Directory | News Releases | Calendar | Articles | | Contact | |
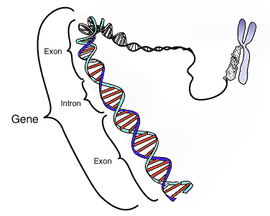
Intron
An intron is a DNA region within a gene that is not translated into protein. These non-coding sections are transcribed to precursor mRNA (pre-mRNA) and some other RNAs (such as long noncoding RNAs), and subsequently removed by a process called splicing during the processing to mature RNA. After intron splicing (ie. removal), the mRNA consists only of exon derived sequences, which are translated into a protein.
The word intron is derived from the term intragenic region and also called intervening sequence (IVS)[1]
Contents |
[edit] Introduction
Introns are common in eukaryotic pre-mRNA, but in prokaryotes they are only found in tRNA and rRNA; introns have variable length and alternate with exons in intron-containing genes.
The number and length of introns varies widely among species, and among genes within the same species. Some eukaryotes, e.g. sac fungi, have evolved genomes with few introns,[2] while the genomes of many other eukaryote groups are rich in introns (several per gene).[3][4]
Alternative splicing of introns within a gene may introduce greater variability of protein sequences translated from a single gene. The control of mRNA splicing is performed by a wide variety of signaling molecules.
Introns may also contain "old code", or sections of a gene that were once translated into a protein, but have since become inactive. It was generally assumed that the sequence of any given intron is junk DNA with no biological function. More recently, however, this is being disputed.[5] For example, a point mutation in intron 7 of the human gene TPH1 is highly correlated to the development of the psychiatric disorder schizophrenia.[6]
Introns contain several short sequences that are important for efficient splicing, such as acceptor and donor sites at either end of the intron as well as a branch point site, which are required for proper splicing by the spliceosome. Some introns are known to enhance the expression of the gene that they are contained in by a process known as intron-mediated enhancement (IME).
One of the most important roles of introns currently under investigation is the transcription of the introns to small regulatory RNA, such as a type of RNAs called miRNA (microRNA). These small single-stranded RNAs regulate the expression of genes.[7]
[edit] Discovery
The discovery of introns led to the Nobel Prize in Physiology or Medicine in 1993 for Phillip Allen Sharp and Richard J. Roberts. The term intron was introduced by American biochemist Walter Gilbert:[8]
"The notion of the cistron [...] must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger - which I suggest we call introns (for intragenic regions) - alternating with regions which will be expressed - exons." (Gilbert 1978)
[edit] Classification
Four classes of introns are known to exist:[citation needed]
Some introns, such as the Group I and Group II introns, after transcription possess ribozyme activity, enabling them to catalyze their own splicing out of a primary RNA transcript. These introns are thus self splicing introns and are relatively rare compared to spliceosomal introns. This self-splicing activity was discovered by Thomas Cech, who shared the 1989 Nobel Prize in Chemistry with Sidney Altman for the discovery of the catalytic properties of RNA.
Nuclear or spliceosomal introns are spliced by the spliceosome and a series of snRNAs (small nuclear RNAs). Certain splice signals (or consensus sequences) abet the splicing (or identification) of these introns by the spliceosome.
Group II and III introns are similar and have a conserved secondary structure. A so-called lariat pathway is used in their splicing. They perform functions similar to the spliceosome and may be evolutionarily related to it. Group I introns are the only class of introns whose splicing requires a free guanine nucleoside. They possess a secondary structure different from that of group II and III introns. Many self-splicing introns code for maturases that help with the splicing process, generally only the splicing of the intron that encodes it.[9]
[edit] Intron evolution
There are two competing theories that offer alternative scenarios for the origin and early evolution of spliceosomal introns. Other classes of introns such as self-splicing and tRNA introns are not subject to much debate, but see [10] for the former. These are popularly referred to as the Introns-Early (IE) and the Introns-Late (IL) views.[11]
The IE model, championed by Walter Gilbert,[12] proposes that introns are extremely old and numerously present in the earliest theoretical ancestors of prokaryotes and eukaryotes, the progenotes. In this model, introns were subsequently lost from prokaryotic organisms, allowing them to attain growth efficiency. A central prediction of this theory is that the early introns were mediators that facilitated the recombination of exons that represented the protein domains.[13] This model cannot account for some observed positional variation of introns shared among related genes.[14]
The IL model proposes that introns were recently inserted into originally intron-less contiguous genes after the divergence of eukaryotes and prokaryotes. In this model, introns probably originated from transposable elements. This model is based on the observation that the spliceosomal introns are restricted to eukaryotes alone. However, there is considerable debate over the presence of introns in the early prokaryote-eukaryote ancestors and the subsequent intron loss-gain during eukaryotic evolution.[15] The evolution of introns and of the intron-exon structure may be largely independent of the evolution of coding-sequences.[16]
[edit] Identification
Nearly all eukaryotic nuclear introns begin with the nucleotide sequence GT, and end with AG (the GT-AG rule). These, along with a larger consensus sequence, help direct the splicing machinery to the proper intronic donor and acceptor sites.
At the time of splicing, the intron is composed of RNA, not DNA. Therefore, the beginning of the sequence in RNA is GU, not GT.
[edit] See also
Structure:
Splicing:
Function
Others:
[edit] References
- ^ Mark Lefers. "intron (intervening sequence)". Department of Biochemistry, Molecular Biology, and Cell Biology, Weinberg College of Arts & Sciences, Northwestern University. http://www.biochem.northwestern.edu/holmgren/Glossary/Definitions/Def-I/intron.html. Retrieved 2008-06-17.
- ^ Stajich JE, Dietrich FS, Roy SW (2007). "Comparative genomic analysis of fungal genomes reveals intron-rich ancestors". Genome Biol. 8 (10): R223. doi:10.1186/gb-2007-8-10-r223. PMID 17949488.
- ^ Csurös M, Rogozin IB, Koonin EV (May 2008). "Extremely intron-rich genes in the alveolate ancestors inferred with a flexible maximum-likelihood approach". Mol. Biol. Evol. 25 (5): 903'11. doi:10.1093/molbev/msn039. PMID 18296415.
- ^ Smith DR, Lee RW (2008). "Nucleotide diversity in the mitochondrial and nuclear compartments of Chlamydomonas reinhardtii: investigating the origins of genome architecture". BMC Evol. Biol. 8: 156. doi:10.1186/1471-2148-8-156. PMID 18495022.
- ^ Crosio C, Cecconi F, Mariottini P, Cesareni G, Brenner S, Amaldi F (December 1996). "Fugu intron oversize reveals the presence of U15 snoRNA coding sequences in some introns of the ribosomal protein S3 gene". Genome Res. 6 (12): e15. doi:10.1101/gr.6.12.1227. PMID 8973918. http://www.genome.org/cgi/pmidlookup?view=long&pmid=8973918.
- ^ Allen NC, Bagade S, McQueen MB, et al. (July 2008). "Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database". Nature Genetics 40 (7): 827'34. doi:10.1038/ng.171. PMID 18583979.
- ^ Shi-Lung Lin, Joseph D. Miller, and Shao-Yao Ying. "Intronic MicroRNA (miRNA)". Department of Cell & Neurobiology, Keck School of Medicine, University of Southern California, BMT-403, 1333 San Pablo Street, Los Angeles, CA 90033, USA. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1559912. Retrieved 2006-01-25.
- ^ Gilbert, Walter (1978). "Why genes in pieces". Nature 271 (5645): 501. doi:10.1038/271501a0.
- ^ Doetsch NA, Thompson MD, Hallick RB (January 1998). "A maturase-encoding group III twintron is conserved in deeply rooted euglenoid species: are group III introns the chicken or the egg?". Mol. Biol. Evol. 15 (1): 76'86. PMID 9491607. http://mbe.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=9491607.
- ^ Gogarten JP, Hilario E. "Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements." BMC Evol Biol. 2006 Nov 13; 6: 94. PMID 17101053
- ^ Roy SW, Gilbert W (March 2006). "The evolution of spliceosomal introns: patterns, puzzles and progress" (PDF). Nat. Rev. Genet. 7 (3): 211'21. doi:10.1038/nrg1807. PMID 16485020. http://www.faculty.biol.ttu.edu/densmore/MB06pdfs/Nat.rev.intron%20evol.pdf.
- ^ Fedorov A, Cao X, Saxonov S, de Souza SJ, Roy SW, Gilbert W. "Intron distribution difference for 276 ancient and 131 modern genes suggests the existence of ancient introns." Proc Natl Acad Sci U S A. 2001 Nov 6; 98(23): 13177-82. PMID 11687643.
- ^ Rodriguez-Trelles F, Tarrio R, Ayala, FJ. "Origins and evolution of spliceosomal introns." Annual Review of Genetics 2006; 40: 47-76.
- ^ Rzhetsky A, Ayala FJ. "The enigma of intron origins." Cellular and Molecular Life Sciences 1999; 55(1): 3-6.
- ^ Sverdlov AV, Csuros M, Rogozin IB, Koonin EV. "A glimpse of a putative pre-intron phase of eukaryotic evolution." Trends Genet. 2007 Mar; 23(3): 105-108. PMID 17239982
- ^ Yandell M, Mungall CJ, Smith C, et al. (March 2006). "Large-scale trends in the evolution of gene structures within 11 animal genomes". PLoS Comput. Biol. 2 (3): e15. doi:10.1371/journal.pcbi.0020015. PMID 16518452.
[edit] External links
|
||||||||
|
SOURCES.COM is an online portal and directory for journalists, news media, researchers and anyone seeking experts, spokespersons, and reliable information resources. Use SOURCES.COM to find experts, media contacts, news releases, background information, scientists, officials, speakers, newsmakers, spokespeople, talk show guests, story ideas, research studies, databases, universities, associations and NGOs, businesses, government spokespeople. Indexing and search applications by Ulli Diemer and Chris DeFreitas.
For information about being included in SOURCES as a expert or spokesperson see the FAQ . For partnerships, content and applications, and domain name opportunities contact us.