| Home | Sources Directory | News Releases | Calendar | Articles | | Contact | |
Amino acid

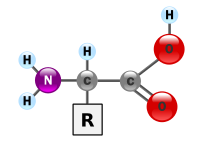
Amino acids are molecules containing an amine group, a carboxylic acid group and a side chain that varies between different amino acids. These molecules contain the key elements of carbon, hydrogen, oxygen, and nitrogen. These molecules are particularly important in biochemistry, where this term usually refers to alpha-amino acids with the general formula H2NCHRCOOH, where R is an organic substituent.[1] In an alpha amino acid, the amino group is attached to the carbon atom immediately adjacent to the carboxylate group (the î�'carbon), but note that other types of amino acid exist when the amino group is attached to another carbon atom (for example, in gamma-amino acids such as gamma-amino-butyric acid the carbon atom to which the amino group attaches is separated from the carboxylate group by two other carbon atoms). The various alpha amino acids differ in which side chain (R group) is attached to their alpha carbon and these can vary in size from just one hydrogen atom in glycine or a methyl group in alanine, through to a large heterocyclic group in tryptophan.
Amino acids are critical to life, and have many functions in metabolism. One particularly important function is to serve as the building blocks of proteins, which are just linear chains of amino acids. Every protein is chemically defined by the order of amino acid residues, their primary structure and this, in turn, determines their secondary structure (for example well-described features such as alpha-helices or beta-pleated sheets), tertiary structure (the shape of the individual protein, for example globular as in most enzymes or linear as in collagen) or quaternary structure (the overall shape of the protein complex if several protein monomers are grouped together, as in the hemoglobin complex, which consists of four hemoglobin monomers held together by hydrogen bonds that are capable of reacting together allosterically when one or more of them binds another molecule for example oxygen). Just as the letters of the alphabet can be combined to form an almost endless variety of words, amino acids can be linked together in varying sequences to form a vast variety of proteins.[2] Amino acids are also important in many other biological molecules, for example they play vital roles in coenzymes such as S-adenosylmethionine. Due to their central role in biochemistry, amino acids are very important in nutrition and are commonly used in food technology and industry. For example, monosodium glutamate is a common flavor enhancer that gives foods the taste called umami. They are also used in industry where applications include the production of biodegradable plastics, drugs and chiral catalysts.
Contents |
[edit] History
The first few amino acids were discovered in the early 1800s. In 1806, the French chemists Louis-Nicolas Vauquelin and Pierre Jean Robiquet isolated a compound in asparagus that proved to be asparagine, the first amino acid to be discovered.[3][4] Another amino acid that was discovered in the early 19th century was cystine, in 1810,[5] although its monomer, cysteine, was discovered much later, in 1884.[4][6] Glycine and leucine were also discovered around this time, in 1820.[7] Usage of the term amino acid in the English language is from 1898.[8]
[edit] General structure
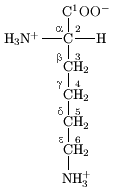
In the structure shown at the top of the page, R represents a side chain specific to each amino acid. The carbon atom next to the carbonyl group is called the î�'carbon and amino acids with a side chain bonded to this carbon are referred to as alpha amino acids. These are the most common form found in nature. In the alpha amino acids, the î�'carbon is a chiral carbon atom, with the exception of glycine.[9] In amino acids that have a carbon chain attached to the î�'carbon (such as lysine, shown to the right) the carbons are labeled in order as î�, î�, î�, î�, and so on.[10] In some amino acids, the amine group is attached to the î� or î�-carbon, and these are therefore referred to as beta or gamma amino acids.
Amino acids are usually classified by the properties of their side chain into four groups. The side chain can make an amino acid a weak acid or a weak base, and a hydrophile if the side chain is polar or a hydrophobe if it is nonpolar.[9] The chemical structures of the twenty-two standard amino acids, along with their chemical properties, are described more fully in the article on these proteinogenic amino acids.
The phrase "branched-chain amino acids" or BCAA refers to the amino acids having aliphatic side chains that are non-linear; these are leucine, isoleucine, and valine. Proline is the only proteinogenic amino acid whose side group links to the î�-amino group and, thus, is also the only proteinogenic amino acid containing a secondary amine at this position.[9] Chemically, proline is therefore an imino acid since it lacks a primary amino group,[11] although it is still classed as an amino acid in the current biochemical nomenclature,[12] and may also be called an "N-alkylated alpha-amino acid".[13]
[edit] Isomerism
Of the standard î�-amino acids, all but glycine can exist in either of two optical isomers, called L or D amino acids, which are mirror images of each other (see also Chirality). While L-amino acids represent all of the amino acids found in proteins during translation in the ribosome, D-amino acids are found in some proteins produced by enzyme posttranslational modification after translation and translocation to the endoplasmic reticulum, as in exotic sea-dwelling organisms such as cone snails.[14] They are also abundant components of the peptidoglycan cell walls of bacteria.[15] and D-serine may act as a neurotransmitter in the brain.[16] The L and D convention for amino acid configuration refers not to the optical activity of the amino acid itself, but rather to the optical activity of the isomer of glyceraldehyde from which that amino acid can theoretically be synthesized (D-glyceraldehyde is dextrorotary; L-glyceraldehyde is levorotary). Alternatively, the (S) and (R) designators are used to indicate the absolute stereochemistry. Almost all of the amino acids in proteins are (S) at the î� carbon, with cysteine being (R) and glycine non-chiral.[17] Cysteine is unusual since it has a sulfur atom at the first position in its side-chain, which has a larger atomic mass than the groups attached to the î�-carbon in the other standard amino acids, thus the (R) instead of (S).
[edit] Zwitterions
Amino acids have both amine and carboxylic acid functional groups and are therefore both an acid and a base at the same time.[9] At a certain pH known as the isoelectric point an amino acid has no overall charge, since the number of protonated ammonium groups (positive charges) and deprotonated carboxylate groups (negative charges) are equal.[18] The amino acids all have different isoelectric points. The ions produced at the isoelectric point have both positive and negative charges and are known as a zwitterion, which comes from the German word Zwitter meaning "hermaphrodite" or "hybrid".[19] Amino acids can exist as zwitterions in solids and in polar solutions such as water, but not in the gas phase.[20] Zwitterions have minimal solubility at their isolectric point and an amino acid can be isolated by precipitating it from water by adjusting the pH to its particular isoelectric point.
[edit] Occurrence and functions in biochemistry
[edit] Standard amino acids
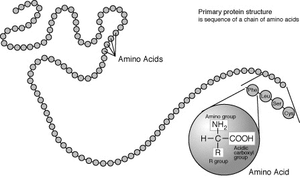
Amino acids are the structural units that make up proteins. They join together to form short polymer chains called peptides or longer chains called either polypeptides or proteins. These polymers are linear and unbranched, with each amino acid within the chain attached to two neighboring amino acids. The process of making proteins is called translation and involves the step-by-step addition of amino acids to a growing protein chain by a ribozyme that is called a ribosome.[21] The order in which the amino acids are added is read through the genetic code from an mRNA template, which is a RNA copy of one of the organism's genes.
Twenty-two amino acids are naturally incorporated into polypeptides and are called proteinogenic or standard amino acids.[9] Of these twenty-two, twenty are directly encoded by the universal genetic code. The remaining two, selenocysteine and pyrrolysine, are incorporated into proteins by unique synthetic mechanisms. Selenocysteine is incorporated when the mRNA being translated includes a SECIS element, which causes the UGA codon to encode selenocysteine instead of a stop codon.[22] Pyrrolysine is used by some methanogenic archaea in enzymes that they use to produce methane. It is coded for with the codon UAG, which is normally a stop codon in other organisms.[23]
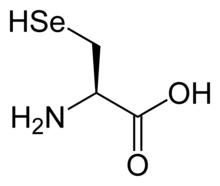
[edit] Non-standard amino acids
Aside from the twenty-two standard amino acids, there are a vast number of "non-standard" amino acids. These non-standard amino acids found in proteins are formed by post-translational modification, which is modification after translation in protein synthesis. These modifications are often essential for the function or regulation of a protein; for example, the carboxylation of glutamate allows for better binding of calcium cations,[24] and the hydroxylation of proline is critical for maintaining connective tissues.[25] Another example is the formation of hypusine in the translation initiation factor EIF5A, through modification of a lysine residue.[26] Such modifications can also determine the localization of the protein, e.g., the addition of long hydrophobic groups can cause a protein to bind to a phospholipid membrane.[27]
Examples of nonstandard amino acids that are not found in proteins include lanthionine, 2-aminoisobutyric acid, dehydroalanine and the neurotransmitter gamma-aminobutyric acid. Nonstandard amino acids often occur as intermediates in the metabolic pathways for standard amino acids ' for example ornithine and citrulline occur in the urea cycle, part of amino acid catabolism (see below).[28] A rare exception to the dominance of î�-amino acids in biology is the î�-amino acid beta alanine (3-aminopropanoic acid), which is used in plants and microorganisms in the synthesis of pantothenic acid (vitamin B5), a component of coenzyme A.[29]
[edit] In human nutrition
When taken up into the human body from the diet, the twenty two standard amino acids are either used to synthesize proteins and other biomolecules or oxidized to urea and carbon dioxide as a source of energy.[30] The oxidation pathway starts with the removal of the amino group by a transaminase, the amino group is then fed into the urea cycle. The other product of transamidation is a keto acid that enters the citric acid cycle.[31] Glucogenic amino acids can also be converted into glucose, through gluconeogenesis.[32]
Pyrrolysine trait is restricted to several microbes, and only one organism has both Pyl and Sec. Of the twenty-two standard amino acids, eight are called essential amino acids because the human body cannot synthesize them from other compounds at the level needed for normal growth, so they must be obtained from food.[33] However, the situation is quite complicated since cysteine, taurine, tyrosine, histidine and arginine are semiessential amino acids in children, because the metabolic pathways that synthesize these amino acids are not fully developed.[34][35] The amounts required also depend on the age and health of the individual, so it is hard to make general statements about the dietary requirement for some amino acids.
| Essential | Nonessential |
|---|---|
| Isoleucine | Alanine |
| Leucine | Asparagine |
| Lysine | Aspartic Acid |
| Methionine | Cysteine* |
| Phenylalanine | Glutamic Acid |
| Threonine | Glutamine* |
| Tryptophan | Glycine* |
| Valine | Proline* |
| Selenocysteine* | |
| Serine* | |
| Tyrosine* | |
| Arginine* | |
| Histidine* | |
| Ornithine* | |
| Taurine* |
(*) Essential only in certain cases.[36][37]
[edit] Non-protein functions
In humans, non-protein amino acids also have important roles as metabolic intermediates, such as in the biosynthesis of the neurotransmitter gamma-aminobutyric acid. Many amino acids are used to synthesize other molecules, for example:
- Tryptophan is a precursor of the neurotransmitter serotonin.[38]
- Glycine is a precursor of porphyrins such as heme.[39]
- Arginine is a precursor of nitric oxide.[40]
- Ornithine and S-adenosylmethionine are precursors of polyamines.[41]
- Aspartate, glycine and glutamine are precursors of nucleotides.[42]
- Phenylalanine is a precursor of various phenylpropanoids which are important in plant metabolism.
However, not all of the functions of other abundant non-standard amino acids are known, for example taurine is a major amino acid in muscle and brain tissues, but although many functions have been proposed, its precise role in the body has not been determined.[43]
Some non-standard amino acids are used as defenses against herbivores in plants.[44] For example canavanine is an analogue of arginine that is found in many legumes,[45] and in particularly large amounts in Canavalia gladiata (sword bean).[46] This amino acid protects the plants from predators such as insects and can cause illness in people if some types of legumes are eaten without processing.[47] The non-protein amino acid mimosine is found in other species of legume, particularly Leucaena leucocephala.[48] This compound is an analogue of tyrosine and can poison animals that graze on these plants.
[edit] Uses in technology
Amino acids are used for a variety of applications in industry but their main use is as additives to animal feed. This is necessary since many of the bulk components of these feeds, such as soybeans, either have low levels or lack some of the essential amino acids: lysine, methionine, threonine, and tryptophan are most important in the production of these feeds.[49] The food industry is also a major consumer of amino acids, particularly glutamic acid, which is used as a flavor enhancer,[50] and Aspartame (aspartyl-phenylalanine-1-methyl ester) as a low-calorie artificial sweetener.[51] The remaining production of amino acids is used in the synthesis of drugs and cosmetics.[49]
| Amino acid derivative | Pharmaceutical application |
|---|---|
| 5-HTP (5-hydroxytryptophan) | Experimental treatment for depression.[52] |
| L-DOPA (L-dihydroxyphenylalanine) | Treatment for Parkinsonism.[53] |
| Eflornithine | Drug that inhibits ornithine decarboxylase and is used in the treatment of sleeping sickness.[54] |
[edit] Expanded genetic code
Since 2001, 40 non-natural amino acids have been added into protein by creating a unique codon (recoding) and a corresponding transfer-RNA:aminoacyl ' tRNA-synthetase pair to encode it with diverse physicochemical and biological properties in order to be used as a tool to exploring protein structure and function or to create novel or enhanced proteins.[55][56]
[edit] Chemical building blocks
Amino acids are important as low-cost feedstocks. These compounds are used in chiral pool synthesis as enantiomerically-pure building blocks.[57]
Amino acids have been investigated as precursors chiral catalysts, e.g. for asymmetric hydrogenation reactions, although no commercial applications exist.[58]
[edit] Biodegradable plastics
Amino acids are under development as components of a range of biodegradable polymers. These materials have applications as environmentally-friendly packaging and in medicine in drug delivery and the construction of prosthetic implants. These polymers include polypeptides, polyamides, polyesters, polysulfides and polyurethanes with amino acids either forming part of their main chains or bonded as side chains. These modifications alter the physical properties and reactivities of the polymers.[59] An interesting example of such materials is polyaspartate, a water-soluble biodegradable polymer that may have applications in disposable diapers and agriculture.[60] Due to its solubility and ability to chelate metal ions, polyaspartate is also being used as a biodegradeable anti-scaling agent and a corrosion inhibitor.[61][62] In addition, the aromatic amino acid tyrosine is being developed as a possible replacement for toxic phenols such as bisphenol A in the manufacture of polycarbonates.[63]
[edit] Reactions
As amino acids have both a primary amine group and a primary carboxyl group, these chemicals can undergo most of the reactions associated with these functional groups. These include nucleophilic addition, amide bond formation and imine formation for the amine group and esterification, amide bond formation and decarboxylation for the carboxylic acid group.[64] The multiple side chains of amino acids can also undergo chemical reactions.[65] The types of these reactions are determined by the groups on these side chains and are therefore different between the various types of amino acid.
[edit] Chemical synthesis
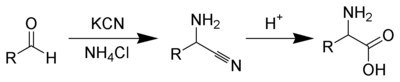
Several methods exist to synthesize amino acids. One of the oldest methods, begins with the bromination at the î�-carbon of a carboxyic acid. Nucleophilic substitution with ammonia then converts the alkyl bromide to the amino acid.[66] Alternatively, the Strecker amino acid synthesis involves the treatment of an aldehyde with potassium cyanide and ammonia, this produces an î�-amino nitrile as an intermediate. Hydrolysis of the nitrile in acid then yields a î�-amino acid.[67] Using ammonia or ammonium salts in this reaction gives unsubstituted amino acids, while substituting primary and secondary amines will yield substituted amino acids.[68] Likewise, using ketones, instead of aldehydes, gives î�,î�-disubstituted amino acids.[69] The classical synthesis gives racemic mixtures of î�-amino acids as products, but several alternative procedures using asymmetric auxiliaries [70] or asymmetric catalysts [71][72] have been developed.[73]
Currently the most adopted method is an automated synthesis on a solid support (e.g. polystyrene beads), using protecting groups (e.g. Fmoc and t-Boc) and activating groups (e.g. DCC and DIC).
[edit] Peptide bond formation
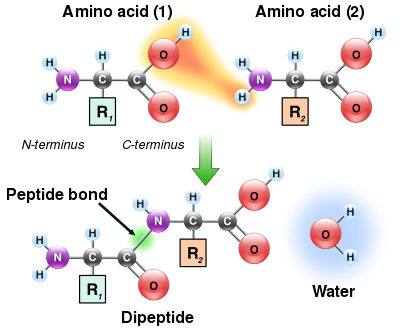
As both the amine and carboxylic acid groups of amino acids can react to form amide bonds, one amino acid molecule can react with another and become joined through an amide linkage. This polymerization of amino acids is what creates proteins. This condensation reaction yields the newly formed peptide bond and a molecule of water. In cells, this reaction does not occur directly; instead the amino acid is first activated by attachment to a transfer RNA molecule through an ester bond. This aminoacyl-tRNA is produced in an ATP-dependent reaction carried out by an aminoacyl tRNA synthetase.[74] This aminoacyl-tRNA is then a substrate for the ribosome, which catalyzes the attack of the amino group of the elongating protein chain on the ester bond.[75] As a result of this mechanism, all proteins made by ribosomes are synthesized starting at their N-terminus and moving towards their C-terminus.
However, not all peptide bonds are formed in this way. In a few cases, peptides are synthesized by specific enzymes. For example, the tripeptide glutathione is an essential part of the defenses of cells against oxidative stress. This peptide is synthesized in two steps from free amino acids.[76] In the first step gamma-glutamylcysteine synthetase condenses cysteine and glutamic acid through a peptide bond formed between the side-chain carboxyl of the glutamate (the gamma carbon of this side chain) and the amino group of the cysteine. This dipeptide is then condensed with glycine by glutathione synthetase to form glutathione.[77]
In chemistry, peptides are synthesized by a variety of reactions. One of the most used in solid-phase peptide synthesis, which uses the aromatic oxime derivatives of amino acids as activated units. These are added in sequence onto the growing peptide chain, which is attached to a solid resin support.[78] The ability to easily synthesize vast numbers of different peptides by varying the types and order of amino acids (using combinatorial chemistry) has made peptide synthesis particularly important in creating libraries of peptides for use in drug discovery through high-throughput screening.[79]
[edit] Biosynthesis and catabolism
In plants, nitrogen is first assimilated into organic compounds in the form of glutamate, formed from alpha-ketoglutarate and ammonia in the mitochondrion. In order to form other amino acids, the plant uses transaminases to move the amino group to another alpha-keto carboxylic acid. For example, aspartate aminotransferase converts glutamate and oxaloacetate to alpha-ketoglutarate and aspartate.[80] Other organisms use transaminases for amino acid synthesis too. Transaminases are also involved in breaking down amino acids. Degrading an amino acid often involves moving its amino group to alpha-ketoglutarate, forming glutamate. In many vertebrates, the amino group is then removed through the urea cycle and is excreted in the form of urea. However, amino acid degradation can produce uric acid or ammonia instead. For example, serine dehydratase converts serine to pyruvate and ammonia.[81]
Nonstandard amino acids are usually formed through modifications to standard amino acids. For example, homocysteine is formed through the transsulfuration pathway or by the demethylation of methionine via the intermediate metabolite S-adenosyl methionine,[43] while hydroxyproline is made by a posttranslational modification of proline.[82]
Microorganisms and plants can synthesize many uncommon amino acids. For example, some microbes make 2-aminoisobutyric acid and lanthionine, which is a sulfide-bridged derivative of alanine. Both of these amino acids are found in peptidic lantibiotics such as alamethicin.[83] While in plants, 1-aminocyclopropane-1-carboxylic acid is a small disubstituted cyclic amino acid that is a key intermediate in the production of the plant hormone ethylene.[84]
[edit] Physicochemical properties of amino acids
The 20 naturally occurring amino acids can be divided into several groups based on their properties. Important factors are charge, hydrophilicity or hydrophobicity, size and functional groups.[9] These properties are important for protein structure and protein'protein interactions. The water-soluble proteins tend to have their hydrophobic residues (Leu, Ile, Val, Phe and Trp) buried in the middle of the protein, whereas hydrophilic side chains are exposed to the aqueous solvent. The integral membrane proteins tend to have outer rings of exposed hydrophobic amino acids that anchor them into the lipid bilayer. In the case part-way between these two extremes, some peripheral membrane proteins have a patch of hydrophobic amino acids on their surface that locks onto the membrane. Similarly, proteins that have to bind to positively-charged molecules have surfaces rich with negatively charged amino acids like glutamate and aspartate, while proteins binding to negatively-charged molecules have surfaces rich with positively charged chains like lysine and arginine. There are different hydrophobicity scales of amino acid residues.[85]
Some amino acids have special properties such as cysteine, that can form covalent disulfide bonds to other cysteine residues, proline that forms a cycle to the polypeptide backbone, and glycine that is more flexible than other amino acids.
Many proteins undergo a range of posttranslational modifications, when additional chemical groups are attached to the amino acids in proteins. Some modifications can produce hydrophobic lipoproteins,[86] or hydrophilic glycoproteins.[87] These type of modification allow the reversible targeting of a protein to a membrane. For example, the addition and removal of the fatty acid palmitic acid to cysteine residues in some signaling proteins causes the proteins to attach and then detach from cell membranes.[88]
[edit] Table of standard amino acid abbreviations and properties
| Amino Acid | 3-Letter[89] | 1-Letter[89] | Side chain polarity[89] | Side chain charge (pH 7.4)[89] | Hydropathy index[90] | Absorbance î»max(nm)[91] | î� at î»max (x10'3 M'1 cm'1)[91] |
|---|---|---|---|---|---|---|---|
| Alanine | Ala | A | nonpolar | neutral | 1.8 | ||
| Arginine | Arg | R | polar | positive | '4.5 | ||
| Asparagine | Asn | N | polar | neutral | '3.5 | ||
| Aspartic acid | Asp | D | polar | negative | '3.5 | ||
| Cysteine | Cys | C | nonpolar | neutral | 2.5 | 250 | 0.3 |
| Glutamic acid | Glu | E | polar | negative | '3.5 | ||
| Glutamine | Gln | Q | polar | neutral | '3.5 | ||
| Glycine | Gly | G | nonpolar | neutral | '0.4 | ||
| Histidine | His | H | polar | positive(10%)
neutral(90%) |
'3.2 | 211 | 5.9 |
| Isoleucine | Ile | I | nonpolar | neutral | 4.5 | ||
| Leucine | Leu | L | nonpolar | neutral | 3.8 | ||
| Lysine | Lys | K | polar | positive | '3.9 | ||
| Methionine | Met | M | nonpolar | neutral | 1.9 | ||
| Phenylalanine | Phe | F | nonpolar | neutral | 2.8 | 257, 206, 188 | 0.2, 9.3, 60.0 |
| Proline | Pro | P | nonpolar | neutral | '1.6 | ||
| Serine | Ser | S | polar | neutral | '0.8 | ||
| Threonine | Thr | T | polar | neutral | '0.7 | ||
| Tryptophan | Trp | W | nonpolar | neutral | '0.9 | 280, 219 | 5.6, 47.0 |
| Tyrosine | Tyr | Y | polar | neutral | '1.3 | 274, 222, 193 | 1.4, 8.0, 48.0 |
| Valine | Val | V | nonpolar | neutral | 4.2 |
Additionally, there are two additional amino acids which are incorporated by overriding stop codons:
| 21st and 22nd amino acids | 3-Letter | 1-Letter |
|---|---|---|
| Selenocysteine | Sec | U |
| Pyrrolysine | Pyl | O |
In addition to the specific amino acid codes, placeholders are used in cases where chemical or crystallographic analysis of a peptide or protein can not conclusively determine the identity of a residue.
| Ambiguous Amino Acids | 3-Letter | 1-Letter |
|---|---|---|
| Asparagine or aspartic acid | Asx | B |
| Glutamine or glutamic acid | Glx | Z |
| Leucine or Isoleucine | Xle | J |
| Unspecified or unknown amino acid | Xaa | X |
Unk is sometimes used instead of Xaa, but is less standard.
Additionally, many non-standard amino acids have a specific code. For example, several peptide drugs, such as Bortezomib or MG132 are artificially synthesized and retain their protecting groups, which have specific codes. Bortezomib is Pyz-Phe-boroLeu and MG132 is Z-Leu-Leu-Leu-al. Additionally, To aid in the analysis of protein structure, photocrosslinking amino acid analogues are available. These include photoleucine (pLeu) and photomethionine (pMet).[92]
[edit] See also
- Amino acid dating
- Beta amino acid
- Degron
- Glucogenic amino acid
- Homochirality
- Leucines
- Miller'Urey experiment
- Proteinogenic amino acid (including chemical structures)
- Table of codons, 3-nucleotide sequences that encode each amino acid
- Polyamide
[edit] References and notes
- ^ Proline is an exception to this general formula. It lacks the NH2 group because of the cyclization of the side chain and is known as an imino acid; it falls under the category of special structured amino acids.
- ^ "The Structures of Life". National Institute of General Medical Sciences. http://publications.nigms.nih.gov/structlife/chapter1.html. Retrieved 2008-05-20.
- ^ Vauquelin LN, Robiquet PJ (1806). "The discovery of a new plant principle in Asparagus sativus". Annales de Chimie 57: 88'93.
- ^ a b Anfinsen CB, Edsall JT, Richards FM (1972). Advances in Protein Chemistry. New York: Academic Press. pp. 99, 103. ISBN 978-0-12-034226-6.
- ^ Wollaston WH (1810). "On cystic oxide, a new species of urinary calculus". Philosophical Transactions of the Royal Society of London 100: 223'30. doi:10.1098/rstl.1810.0015.
- ^ Baumann E (1884). "Über cystin und cystein". Z Physiol Chemie 8: 299.
- ^ Braconnot HM (1820). "Sur la conversion des matières animales en nouvelles substances par le moyen de l'acide sulfurique". Ann Chim Phys Ser 2 13: 113'25.
- ^ "etymonline.com entry for amino". www.etymonline.com. http://www.etymonline.com/index.php?term=amino. Retrieved 2010-07-19.
- ^ a b c d e f Creighton, Thomas H. (1993). "Chapter 1". Proteins: structures and molecular properties. San Francisco: W. H. Freeman. ISBN 978-0-7167-7030-5.
- ^ "Nomenclature and Symbolism for Amino Acids and Peptides". IUPAC-IUB Joint Commission on Biochemical Nomenclature. 1983. http://www.chem.qmul.ac.uk/iupac/AminoAcid/AA1n2.html. Retrieved 2008-11-17.
- ^ Jodidi, S. L. (1926-03-01). "The Formol Titration of Certain Amino Acids". Journal of the American Chemical Society 48 (3): 751'753. doi:10.1021/ja01414a033.
- ^ Liebecq, Claude, ed (1992). Biochemical Nomenclature and Related Documents (2nd ed.). Portland Press. pp. 39'69. ISBN 978-1-85578-005-7.
- ^ Smith, Anthony D. (1997). Oxford dictionary of biochemistry and molecular biology. Oxford: Oxford University Press. pp. 535. ISBN 978-0-19-854768-6. OCLC 37616711.
- ^ Pisarewicz K, Mora D, Pflueger FC, Fields GB, Marí F (May 2005). "Polypeptide chains containing D-gamma-hydroxyvaline". Journal of the American Chemical Society 127 (17): 6207'15. doi:10.1021/ja050088m. PMID 15853325.
- ^ van Heijenoort J (March 2001). "Formation of the glycan chains in the synthesis of bacterial peptidoglycan". Glycobiology 11 (3): 25R'36R. doi:10.1093/glycob/11.3.25R. PMID 11320055.
- ^ Wolosker H, Dumin E, Balan L, Foltyn VN (July 2008). "D-amino acids in the brain: D-serine in neurotransmission and neurodegeneration". The FEBS Journal 275 (14): 3514'26. doi:10.1111/j.1742-4658.2008.06515.x. PMID 18564180.
- ^ Hatem, Salama Mohamed Ali (2006). "Gas chromatographic determination of Amino Acid Enantiomers in tobacco and bottled wines". University of Giessen. http://geb.uni-giessen.de/geb/volltexte/2006/3038/index.html. Retrieved 2008-11-17.
- ^ Fennema OR (1996). Food Chemistry (3rd ed.). CRC Press. pp. 327'8. ISBN 0-8247-9691-8.
- ^ Simmons, William J.; Gerhard Meisenberg (2006). Principles of medical biochemistry. Mosby Elsevier. p. 19. ISBN 0-323-02942-6.
- ^ Remko M, Rode BM (February 2006). "Effect of metal ions (Li+, Na+, K+, Mg2+, Ca2+, Ni2+, Cu2+, and Zn2+) and water coordination on the structure of glycine and zwitterionic glycine". The journal of physical chemistry. A 110 (5): 1960'7. doi:10.1021/jp054119b. PMID 16451030.
- ^ Rodnina MV, Beringer M, Wintermeyer W (January 2007). "How ribosomes make peptide bonds". Trends in Biochemical Sciences 32 (1): 20'6. doi:10.1016/j.tibs.2006.11.007. PMID 17157507.
- ^ Driscoll DM, Copeland PR (2003). "Mechanism and regulation of selenoprotein synthesis". Annual Review of Nutrition 23: 17'40. doi:10.1146/annurev.nutr.23.011702.073318. PMID 12524431.
- ^ Krzycki JA (December 2005). "The direct genetic encoding of pyrrolysine". Current Opinion in Microbiology 8 (6): 706'12. doi:10.1016/j.mib.2005.10.009. PMID 16256420.
- ^ Vermeer C (March 1990). "Gamma-carboxyglutamate-containing proteins and the vitamin K-dependent carboxylase". The Biochemical Journal 266 (3): 625'36. PMID 2183788.
- ^ Bhattacharjee A, Bansal M (March 2005). "Collagen structure: the Madras triple helix and the current scenario". IUBMB Life 57 (3): 161'72. doi:10.1080/15216540500090710. PMID 16036578.
- ^ Park MH (February 2006). "The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A)". Journal of Biochemistry 139 (2): 161'9. doi:10.1093/jb/mvj034. PMID 16452303.
- ^ Blenis J, Resh MD (December 1993). "Subcellular localization specified by protein acylation and phosphorylation". Current Opinion in Cell Biology 5 (6): 984'9. doi:10.1016/0955-0674(93)90081-Z. PMID 8129952.
- ^ Curis E, Nicolis I, Moinard C, et al. (November 2005). "Almost all about citrulline in mammals". Amino Acids 29 (3): 177'205. doi:10.1007/s00726-005-0235-4. PMID 16082501.
- ^ Coxon KM, Chakauya E, Ottenhof HH, et al. (August 2005). "Pantothenate biosynthesis in higher plants". Biochemical Society Transactions 33 (Pt 4): 743'6. doi:10.1042/BST0330743. PMID 16042590.
- ^ Sakami W, Harrington H (1963). "Amino acid metabolism". Annual Review of Biochemistry 32: 355'98. doi:10.1146/annurev.bi.32.070163.002035. PMID 14144484.
- ^ Brosnan JT (April 2000). "Glutamate, at the interface between amino acid and carbohydrate metabolism". The Journal of Nutrition 130 (4S Suppl): 988S'90S. PMID 10736367. http://jn.nutrition.org/cgi/pmidlookup?view=long&pmid=10736367.
- ^ Young VR, Ajami AM (September 2001). "Glutamine: the emperor or his clothes?". The Journal of Nutrition 131 (9 Suppl): 2449S'59S; discussion 2486S'7S. PMID 11533293. http://jn.nutrition.org/cgi/pmidlookup?view=long&pmid=11533293.
- ^ Young VR (August 1994). "Adult amino acid requirements: the case for a major revision in current recommendations". The Journal of Nutrition 124 (8 Suppl): 1517S'1523S. PMID 8064412. http://jn.nutrition.org/cgi/pmidlookup?view=long&pmid=8064412.
- ^ Imura K, Okada A (January 1998). "Amino acid metabolism in pediatric patients". Nutrition 14 (1): 143'8. doi:10.1016/S0899-9007(97)00230-X. PMID 9437700.
- ^ Lourenço R, Camilo ME (2002). "Taurine: a conditionally essential amino acid in humans? An overview in health and disease". Nutrición Hospitalaria 17 (6): 262'70. PMID 12514918.
- ^ Fürst P, Stehle P (June 2004). "What are the essential elements needed for the determination of amino acid requirements in humans?". The Journal of Nutrition 134 (6 Suppl): 1558S'1565S. PMID 15173430. http://jn.nutrition.org/cgi/pmidlookup?view=long&pmid=15173430.
- ^ Reeds PJ (July 2000). "Dispensable and indispensable amino acids for humans". The Journal of Nutrition 130 (7): 1835S'40S. PMID 10867060. http://jn.nutrition.org/cgi/pmidlookup?view=long&pmid=10867060.
- ^ Savelieva KV, Zhao S, Pogorelov VM, et al. (2008). "Genetic disruption of both tryptophan hydroxylase genes dramatically reduces serotonin and affects behavior in models sensitive to antidepressants". PloS ONE 3 (10): e3301. doi:10.1371/journal.pone.0003301. PMID 18923670.
- ^ Shemin D, Rittenberg D (1 December 1946). "The biological utilization of glycine for the synthesis of the protoporphyrin of hemoglobin". Journal of Biological Chemistry 166 (2): 621'5. PMID 20276176. http://www.jbc.org/cgi/reprint/166/2/621.
- ^ Tejero J, Biswas A, Wang ZQ, et al. (November 2008). "Stabilization and characterization of a heme-oxy reaction intermediate in inducible nitric-oxide synthase". The Journal of Biological Chemistry 283 (48): 33498'507. doi:10.1074/jbc.M806122200. PMID 18815130.
- ^ Rodríguez-Caso C, Montañez R, Cascante M, S�¡nchez-Jiménez F, Medina MA (August 2006). "Mathematical modeling of polyamine metabolism in mammals". The Journal of Biological Chemistry 281 (31): 21799'812. doi:10.1074/jbc.M602756200. PMID 16709566.
- ^ Stryer, Lubert; Berg, Jeremy Mark; Tymoczko, John L. (2002). Biochemistry. San Francisco: W.H. Freeman. pp. 693'8. ISBN 0-7167-4684-0.
- ^ a b Brosnan JT, Brosnan ME (June 2006). "The sulfur-containing amino acids: an overview". The Journal of Nutrition 136 (6 Suppl): 1636S'1640S. PMID 16702333. http://jn.nutrition.org/cgi/pmidlookup?view=long&pmid=16702333.
- ^ Hylin, John W. (1969). "Toxic peptides and amino acids in foods and feeds". Journal of Agricultural and Food Chemistry 17 (3): 492'6. doi:10.1021/jf60163a003.
- ^ Turner, B. L.; Harborne, J. B. (1967). "Distribution of canavanine in the plant kingdom". Phytochemistry 6: 863'66. doi:10.1016/S0031-9422(00)86033-1.
- ^ Ekanayake S, Skog K, Asp NG (May 2007). "Canavanine content in sword beans (Canavalia gladiata): analysis and effect of processing". Food and Chemical Toxicology 45 (5): 797'803. doi:10.1016/j.fct.2006.10.030. PMID 17187914.
- ^ Rosenthal GA (2001). "L-Canavanine: a higher plant insecticidal allelochemical". Amino Acids 21 (3): 319'30. doi:10.1007/s007260170017. PMID 11764412.
- ^ Hammond AC (May 1995). "Leucaena toxicosis and its control in ruminants". Journal of Animal Science 73 (5): 1487'92. PMID 7665380. http://jas.fass.org/cgi/pmidlookup?view=long&pmid=7665380.
- ^ a b Leuchtenberger W, Huthmacher K, Drauz K (November 2005). "Biotechnological production of amino acids and derivatives: current status and prospects". Applied Microbiology and Biotechnology 69 (1): 1'8. doi:10.1007/s00253-005-0155-y. PMID 16195792.
- ^ Garattini S (April 2000). "Glutamic acid, twenty years later". The Journal of Nutrition 130 (4S Suppl): 901S'9S. PMID 10736350. http://jn.nutrition.org/cgi/pmidlookup?view=long&pmid=10736350.
- ^ Stegink LD (July 1987). "The aspartame story: a model for the clinical testing of a food additive". The American Journal of Clinical Nutrition 46 (1 Suppl): 204'15. PMID 3300262. http://www.ajcn.org/cgi/pmidlookup?view=long&pmid=3300262.
- ^ Turner EH, Loftis JM, Blackwell AD (March 2006). "Serotonin a la carte: supplementation with the serotonin precursor 5-hydroxytryptophan". Pharmacology & Therapeutics 109 (3): 325'38. doi:10.1016/j.pharmthera.2005.06.004. PMID 16023217.
- ^ Kostrzewa RM, Nowak P, Kostrzewa JP, Kostrzewa RA, Brus R (March 2005). "Peculiarities of L: -DOPA treatment of Parkinson's disease". Amino Acids 28 (2): 157'64. doi:10.1007/s00726-005-0162-4. PMID 15750845.
- ^ Heby O, Persson L, Rentala M (August 2007). "Targeting the polyamine biosynthetic enzymes: a promising approach to therapy of African sleeping sickness, Chagas' disease, and leishmaniasis". Amino Acids 33 (2): 359'66. doi:10.1007/s00726-007-0537-9. PMID 17610127.
- ^ Xie J, Schultz PG (December 2005). "Adding amino acids to the genetic repertoire". Curr Opin Chem Biol 9 (6): 548'54. doi:10.1016/j.cbpa.2005.10.011. PMID 16260173.
- ^ Wang Q, Parrish AR, Wang L (March 2009). "Expanding the genetic code for biological studies". Chem. Biol. 16 (3): 323'36. doi:10.1016/j.chembiol.2009.03.001. PMID 19318213.
- ^ Hanessian, S. (1993). "Reflections on the total synthesis of natural products: Art, craft, logic, and the chiron approach". Pure and Applied Chemistry 65: 1189'204. doi:10.1351/pac199365061189.
- ^ Blaser, Hans Ulrich (1992). "The chiral pool as a source of enantioselective catalysts and auxiliaries". Chemical Reviews 92 (5): 935'52. doi:10.1021/cr00013a009.
- ^ Sanda, Fumio; Endo, Takeshi (1999). "Feature Article Syntheses and functions of polymers based on amino acids". Macromolecular Chemistry and Physics 200: 2651'61. doi:10.1002/(SICI)1521-3935(19991201)200:12<2651::AID-MACP2651>3.0.CO;2-P.
- ^ Gross, R. A.; Kalra, B. (2002). "Biodegradable Polymers for the Environment". Science 297 (5582): 803'807. doi:10.1126/science.297.5582.803. PMID 12161646. http://www.sciencemag.org/cgi/content/abstract/297/5582/803.
- ^ Low, K. C.; Wheeler, A. P.; Koskan, L. P. (1996). Commercial poly(aspartic acid) and Its Uses. Advances in Chemistry Series. 248. Washington, D.C.: American Chemical Society.
- ^ Thombre, S.M.; Sarwade, B.D. (2005). "Synthesis and Biodegradability of Polyaspartic Acid: A Critical Review". Journal of Macromolecular Science, Part A 42 (9): 1299'1315. doi:10.1080/10601320500189604. http://www.informaworld.com/index/718581646.pdf.
- ^ Bourke, S. L.; Kohn, J. (2003). "Polymers derived from the amino acid l-tyrosine: polycarbonates, polyarylates and copolymers with poly(ethylene glycol)". Advanced Drug Delivery Reviews 55 (4): 447'466. doi:10.1016/S0169-409X(03)00038-3. PMID 12706045. http://linkinghub.elsevier.com/retrieve/pii/S0169409X03000383.
- ^ Elmore, Donald Trevor; Barrett, G. C. (1998). Amino acids and peptides. Cambridge, UK: Cambridge University Press. pp. 48'60. ISBN 0-521-46827-2.
- ^ Gutteridge A, Thornton JM (November 2005). "Understanding nature's catalytic toolkit". Trends in Biochemical Sciences 30 (11): 622'9. doi:10.1016/j.tibs.2005.09.006. PMID 16214343.
- ^ McMurry, John (1996). Organic chemistry. Pacific Grove, CA, USA: Brooks/Cole. p. 1064. ISBN 0-534-23832-7.
- ^ Strecker, Adolph (1850). "Ueber die künstliche Bildung der Milchsäure und einen neuen, dem Glycocoll homologen Körper". Justus Liebigs Annalen der Chemie 75 (1): 27'45. doi:10.1002/jlac.18500750103.
- ^ Strecker, Adolph (1854). "Ueber einen neuen aus Aldehyd - Ammoniak und Blausäure entstehenden Körper". Justus Liebigs Annalen der Chemie 91 (3): 349'51. doi:10.1002/jlac.18540910309.
- ^ Masumoto S, Usuda H, Suzuki M, Kanai M, Shibasaki M (May 2003). "Catalytic enantioselective Strecker reaction of ketoimines". Journal of the American Chemical Society 125 (19): 5634'5. doi:10.1021/ja034980. PMID 12733893.
- ^ Davis, F. A. (1994). Tetrahedron Letters 35: 9351.
- ^ Ishitani, Haruro; Komiyama, Susumu; Hasegawa, Yoshiki; Kobayashi, ShÅ« (2000). "Catalytic Asymmetric Strecker Synthesis. Preparation of Enantiomerically Pure î�-Amino Acid Derivatives from Aldimines and Tributyltin Cyanide or Achiral Aldehydes, Amines, and Hydrogen Cyanide Using a Chiral Zirconium Catalyst". Journal of the American Chemical Society 122 (5): 762'6. doi:10.1021/ja9935207.
- ^ Huang, Jinkun; Corey, E. J. (2004). "A New Chiral Catalyst for the Enantioselective Strecker Synthesis of î�-Amino Acids". Orgic Letters 62 (6): 5027'9. doi:10.1021/ol047698w. PMID 15606127.
- ^ Duthaler, Rudolf O. (1994). "Recent developments in the stereoselective synthesis of î�-aminoacids". Tetrahedron 50 (6): 1539'1650. doi:10.1016/S0040-4020(01)80840-1.
- ^ Ibba M, Söll D (May 2001). "The renaissance of aminoacyl-tRNA synthesis". EMBO Reports 2 (5): 382'7. doi:10.1093/embo-reports/kve095 (inactive 2010-02-18). PMID 11375928. PMC 1083889. http://www.nature.com/embor/journal/v2/n5/full/embor420.html.
- ^ Lengyel P, Söll D (June 1969). "Mechanism of protein biosynthesis". Bacteriological Reviews 33 (2): 264'301. PMID 4896351. PMC 378322. http://mmbr.asm.org/cgi/pmidlookup?view=long&pmid=4896351.
- ^ Wu G, Fang YZ, Yang S, Lupton JR, Turner ND (March 2004). "Glutathione metabolism and its implications for health". The Journal of Nutrition 134 (3): 489'92. PMID 14988435. http://jn.nutrition.org/cgi/pmidlookup?view=long&pmid=14988435.
- ^ Meister A (November 1988). "Glutathione metabolism and its selective modification". The Journal of Biological Chemistry 263 (33): 17205'8. PMID 3053703. http://www.jbc.org/cgi/pmidlookup?view=long&pmid=3053703.
- ^ Carpino, Louis A. (1992). "1-Hydroxy-7-azabenzotriazole. An efficient peptide coupling additive". Journal of the American Chemical Society 115 (10): 4397'8. doi:10.1021/ja00063a082.
- ^ Marasco D, Perretta G, Sabatella M, Ruvo M (October 2008). "Past and future perspectives of synthetic peptide libraries". Current Protein & Peptide Science 9 (5): 447'67. doi:10.2174/138920308785915209. PMID 18855697.
- ^ Jones, Russell Celyn; Buchanan, Bob B.; Gruissem, Wilhelm (2000). Biochemistry & molecular biology of plants. Rockville, Md: American Society of Plant Physiologists. pp. 371'2. ISBN 0-943088-39-9.
- ^ Stryer, Lubert; Berg, Jeremy Mark; Tymoczko, John L. (2002). Biochemistry. San Francisco: W.H. Freeman. pp. 639'49. ISBN 0-7167-4684-0.
- ^ Kivirikko KI, Pihlajaniemi T (1998). "Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases". Advances in Enzymology and Related Areas of Molecular Biology 72: 325'98. PMID 9559057.
- ^ Whitmore L, Wallace BA (May 2004). "Analysis of peptaibol sequence composition: implications for in vivo synthesis and channel formation". European Biophysics Journal 33 (3): 233'7. doi:10.1007/s00249-003-0348-1. PMID 14534753.
- ^ Alexander L, Grierson D (October 2002). "Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening". Journal of Experimental Botany 53 (377): 2039'55. doi:10.1093/jxb/erf072. PMID 12324528.
- ^ Urry, Dan W. (2004). "The change in Gibbs free energy for hydrophobic association: Derivation and evaluation by means of inverse temperature transitions". Chemical Physics Letters 399 (1'3): 177'83. doi:10.1016/S0009-2614(04)01565-9.
- ^ Magee T, Seabra MC (April 2005). "Fatty acylation and prenylation of proteins: what's hot in fat". Current Opinion in Cell Biology 17 (2): 190'6. doi:10.1016/j.ceb.2005.02.003. PMID 15780596.
- ^ Pilobello KT, Mahal LK (June 2007). "Deciphering the glycocode: the complexity and analytical challenge of glycomics". Current Opinion in Chemical Biology 11 (3): 300'5. doi:10.1016/j.cbpa.2007.05.002. PMID 17500024.
- ^ Smotrys JE, Linder ME (2004). "Palmitoylation of intracellular signaling proteins: regulation and function". Annual Review of Biochemistry 73: 559'87. doi:10.1146/annurev.biochem.73.011303.073954. PMID 15189153.
- ^ a b c d Hausman, Robert E.; Cooper, Geoffrey M. (2004). The cell: a molecular approach. Washington, D.C: ASM Press. p. 51. ISBN 0-87893-214-3.
- ^ Kyte J, Doolittle RF (May 1982). "A simple method for displaying the hydropathic character of a protein". Journal of Molecular Biology 157 (1): 105'32. doi:10.1016/0022-2836(82)90515-0. PMID 7108955.
- ^ a b Freifelder, D. (1983). Physical Biochemistry (2nd ed.). W. H. Freeman and Company. ISBN 0-7167-1315-2.[page needed]
- ^ Suchanek M, Radzikowska A, Thiele C (April 2005). "Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells". Nature Methods 2 (4): 261'7. doi:10.1038/nmeth752. PMID 15782218.
[edit] Further reading
- Doolittle, R.F. (1989) Redundancies in protein sequences. In Predictions of Protein Structure and the Principles of Protein Conformation (Fasman, G.D. ed) Plenum Press, New York, pp. 599'623
- David L. Nelson and Michael M. Cox, Lehninger Principles of Biochemistry, 3rd edition, 2000, Worth Publishers, ISBN 1-57259-153-6
- Meierhenrich, U.J.: Amino acids and the asymmetry of life, Springer-Verlag, Berlin, New York, 2008. ISBN 978-3-540-76885-2
- Morelli, Robert J. "Studies of amino acid absorption from the small intestine." San Francisco: Morelli, 1952.
[edit] External links
- Amino acids overview physical-chemistry properties, 3D structures, etc
- List of Standard Amino Acids The Detailed PDF List of Standard Amino Acids (including 3D depictions)
- Nomenclature and Symbolism for Amino Acids and Peptides IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN)
- Molecular Expressions: The Amino Acid Collection ' Has detailed information and microscopy photographs of each amino acid.
- Amino acid properties ' Properties of the amino acids (a tool aimed mostly at molecular geneticists trying to understand the meaning of mutations)
- Synthesis of Amino Acids and Derivatives
- Learn the 20 proteinogenic amino acids online
- The origin of the single-letter code for the amino acids
- Amino acid solution's pH, titration and isoelectric point calculation free spreadsheet
|
||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
SOURCES.COM is an online portal and directory for journalists, news media, researchers and anyone seeking experts, spokespersons, and reliable information resources. Use SOURCES.COM to find experts, media contacts, news releases, background information, scientists, officials, speakers, newsmakers, spokespeople, talk show guests, story ideas, research studies, databases, universities, associations and NGOs, businesses, government spokespeople. Indexing and search applications by Ulli Diemer and Chris DeFreitas.
For information about being included in SOURCES as a expert or spokesperson see the FAQ . For partnerships, content and applications, and domain name opportunities contact us.