| Home | Sources Directory | News Releases | Calendar | Articles | | Contact | |
Apoptosis
Apoptosis (pronounced /ˌæpÉ�ˈtoÊŠsÉ�s/ or /ˌæpÉ�pˈtoÊŠsÉ�s/)[1][2] is the process of programmed cell death (PCD) that may occur in multicellular organisms. Biochemical events lead to characteristic cell changes (morphology) and death. These changes include blebbing, loss of cell membrane asymmetry and attachment, cell shrinkage, nuclear fragmentation, chromatin condensation, and chromosomal DNA fragmentation. (See also Apoptosis DNA fragmentation.) Apoptosis differs from necrosis, in which the cellular debris can damage the organism.
In contrast to necrosis, which is a form of traumatic cell death that results from acute cellular injury, apoptosis, in general, confers advantages during an organism's life cycle. For example, the differentiation of fingers and toes in a developing human embryo occurs because cells between the fingers apoptose; the result is that the digits are separate. Between 50 and 70 billion cells die each day due to apoptosis in the average human adult. For an average child between the ages of 8 and 14, approximately 20 billion to 30 billion cells die a day.
Research in and around apoptosis has increased substantially since the early 1990s. In addition to its importance as a biological phenomenon, defective apoptotic processes have been implicated in an extensive variety of diseases. Excessive apoptosis causes atrophy, such as in ischemic damage, whereas an insufficient amount results in uncontrolled cell proliferation, such as cancer.
Contents |
[edit] Discovery and etymology
German scientist Carl Vogt was first to describe the principle of apoptosis in 1842. In 1885, anatomist Walther Flemming delivered a more precise description of the process of programmed cell death. However, it was not until 1965 that the topic was resurrected. While studying tissues using electron microscopy, John Foxton Ross Kerr at University of Queensland was able to distinguish apoptosis (Greek: apo - from/off/without, ptosis - falling) from traumatic cell death.[3] Following the publication of a paper describing the phenomenon, Kerr was invited to join Alastair R Currie, as well as Andrew Wyllie, who was Currie's graduate student,[4] at University of Aberdeen. In 1972, the trio published a seminal article in the British Journal of Cancer.[5] Kerr had initially used the term programmed cell necrosis, but in the article, the process of natural cell death was called apoptosis. Kerr, Wyllie and Currie credited James Cormack, a professor of Greek language at University of Aberdeen, with suggesting the term apoptosis. Kerr received the Paul Ehrlich and Ludwig Darmstaedter Prize on March 14, 2000, for his description of apoptosis. He shared the prize with Boston biologist Robert Horvitz.[6]. The 2002 Nobel Prize in Medicine was awarded to Sydney Brenner, Horvitz and John E. Sulston for their work regarding apoptosis.
In Greek, apoptosis translates to the "dropping off" of petals or leaves from plants or trees. Cormack, professor of Greek language, reintroduced the term for medical use as it had a medical meaning for the Greeks over two thousand years before. Hippocrates used the term to mean "the falling off of the bones". Galen extended its meaning to "the dropping of the scabs". Cormack was no doubt aware of this usage when he suggested the name. Debate continues over the correct pronunciation, with opinion divided between a pronunciation with the second p silent (pronounced /æpÉ�ˈtoÊŠsÉ�s/ ap-É�-TOE-sis[1][7]) and the second p pronounced (pronounced /æpÉ�pˈtoÊŠsÉ�s/),[1][8] as in the original Greek.[citation needed] In English, the p of the Greek -pt- consonant cluster is typically silent at the beginning of a word (e.g. pterodactyl, Ptolemy), but articulated when used in combining forms preceded by a vowel, as in helicopter or the orders of insects: diptera, lepidoptera, etc.
In the original Kerr Wyllie and Currie paper, British Journal of Cancer, 1972 Aug;26(4):239-57, there is a footnote regarding the pronunciation:
"We are most grateful to Professor James Cormack of the Department of Greek, University of Aberdeen, for suggesting this term. The word "apoptosis" (Greek spelling of apoptosis) is used in Greek to describe the "dropping off" or "falling off" of petals from flowers, or leaves from trees. To show the derivation clearly, we propose that the stress should be on the penultimate syllable, the second half of the word being pronounced like "ptosis" (with the "p" silent), which comes from the same root "to fall", and is already used to describe the drooping of the upper eyelid."
[edit] Process
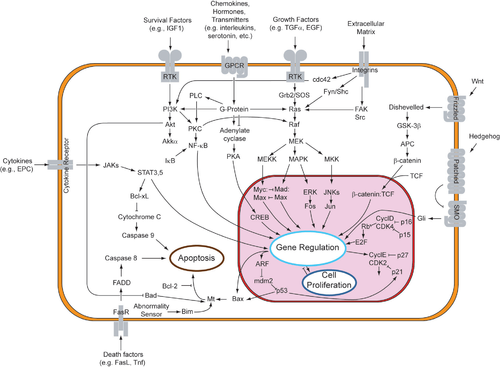
The process of apoptosis is controlled by a diverse range of cell signals, which may originate either extracellularly (extrinsic inducers) or intracellularly (intrinsic inducers). Extracellular signals may include toxins[9], hormones, growth factors, nitric oxide[10] or cytokines, that must either cross the plasma membrane or transduce to effect a response. These signals may positively (i.e., trigger) or negatively (i.e., repress, inhibit, or dampen) affect apoptosis. (Binding and subsequent initiation of apoptosis by a molecule is termed positive induction, whereas the active repression or inhibition of apoptosis by a molecule is termed negative induction.)
A cell initiates intracellular apoptotic signalling in response to a stress, which may bring about cell suicide. The binding of nuclear receptors by glucocorticoids[11], heat[11], radiation[11], nutrient deprivation[11], viral infection[11], hypoxia[11] and increased intracellular calcium concentration[12], for example, by damage to the membrane, can all trigger the release of intracellular apoptotic signals by a damaged cell. A number of cellular components, such as poly ADP ribose polymerase, may also help regulate apoptosis.[13]
Before the actual process of cell death is precipitated by enzymes, apoptotic signals must cause regulatory proteins to initiate the apoptosis pathway. This step allows apoptotic signals to cause cell death, or the process to be stopped, should the cell no longer need to die. Several proteins are involved, but two main methods of regulation have been identified: targeting mitochondria functionality, or directly transducing the signal via adaptor proteins to the apoptotic mechanisms. Another extrinsic pathway for initiation identified in several toxin studies is an increase in calcium concentration within a cell caused by drug activity, which also can cause apoptosis via a calcium binding protease calpain.
[edit] Mitochondrial regulation
The mitochondria are essential to multicellular life. Without them, a cell ceases to respire aerobically and quickly dies, a fact exploited by some apoptotic pathways. Apoptotic proteins that target mitochondria affect them in different ways. They may cause mitochondrial swelling through the formation of membrane pores, or they may increase the permeability of the mitochondrial membrane and cause apoptotic effectors to leak out.[11] There is also a growing body of evidence indicating that nitric oxide is able to induce apoptosis by helping to dissipate the membrane potential of mitochondria and therefore make it more permeable.[10]
Mitochondrial proteins known as SMACs (second mitochondria-derived activator of caspases) are released into the cytosol following an increase in permeability. SMAC binds to inhibitor of apoptosis proteins (IAPs) and deactivates them, preventing the IAPs from arresting the apoptotic process and therefore allowing apoptosis to proceed. IAP also normally suppresses the activity of a group of cysteine proteases called caspases,[14] which carry out the degradation of the cell, therefore the actual degradation enzymes can be seen to be indirectly regulated by mitochondrial permeability.
Cytochrome c is also released from mitochondria due to formation of a channel, MAC, in the outer mitochondrial membrane[15], and serves a regulatory function as it precedes morphological change associated with apoptosis.[11] Once cytochrome c is released it binds with Apoptotic protease activating factor - 1Apaf-1 and ATP, which then bind to pro-caspase-9 to create a protein complex known as an apoptosome. The apoptosome cleaves the pro-caspase to its active form of caspase-9, which in turn activates the effector caspase-3.
MAC, also called "Mitochondrial Outer Permeabilization Pore" is regulated by various proteins, such as those encoded by the mammalian Bcl-2 family of anti-apoptopic genes, the homologs of the ced-9 gene found in C. elegans.[16][17] Bcl-2 proteins are able to promote or inhibit apoptosis by direct action on MAC/MOMP. Bax and/or Bak form the pore, while Bcl-2, Bcl-xL or Mcl-1 inhibit its formation.
[edit] Direct signal transduction
Two theories of the direct initiation of apoptotic mechanisms in mammals have been suggested: the TNF-induced (tumour necrosis factor) model and the Fas-Fas ligand-mediated model, both involving receptors of the TNF receptor (TNFR) family[18] coupled to extrinsic signals.
TNF is a cytokine produced mainly by activated macrophages, and is the major extrinsic mediator of binary hipaloptic apoptosis. Most cells in the human body have two receptors for TNF: TNF-R1 and TNF-R2. The binding of TNF to TNF-R1 has been shown to initiate the pathway that leads to caspase activation via the intermediate membrane proteins TNF receptor-associated death domain (TRADD) and Fas-associated death domain protein (FADD).[19] Binding of this receptor can also indirectly lead to the activation of transcription factors involved in cell survival and inflammatory responses.[20] The link between TNF and apoptosis shows why an abnormal production of TNF plays a fundamental role in several human diseases, especially in autoimmune diseases.
The Fas receptor (also known as Apo-1 or CD95) binds the Fas ligand (FasL), a transmembrane protein part of the TNF family.[18] The interaction between Fas and FasL results in the formation of the death-inducing signaling complex (DISC), which contains the FADD, caspase-8 and caspase-10. In some types of cells (type I), processed caspase-8 directly activates other members of the caspase family, and triggers the execution of apoptosis of the cell. In other types of cells (type II), the Fas-DISC starts a feedback loop that spirals into increasing release of pro-apoptotic factors from mitochondria and the amplified activation of caspase-8.[21]
Following TNF-R1 and Fas activation in mammalian cells a balance between pro-apoptotic (BAX,[22] BID, BAK, or BAD) and anti-apoptotic (Bcl-Xl and Bcl-2) members of the Bcl-2 family is established. This balance is the proportion of pro-apoptotic homodimers that form in the outer-membrane of the mitochondrion. The pro-apoptotic homodimers are required to make the mitochondrial membrane permeable for the release of caspase activators such as cytochrome c and SMAC. Control of pro-apoptotic proteins under normal cell conditions of non-apoptotic cells is incompletely understood, but in general, Bax or Bak are activated by the activation of BH3-only proteins, part of the Bcl-2 family.
There also exists a caspase-independent apoptotic pathway that is mediated by AIF (apoptosis-inducing factor).[23]
[edit] Execution
Many pathways and signals lead to apoptosis, but there is only one mechanism that actually causes the death of a cell.[citation needed] After a cell receives stimulus, it undergoes organized degradation of cellular organelles by activated proteolytic caspases. A cell undergoing apoptosis shows a characteristic morphology:
- Cell shrinkage and rounding are shown because of the breakdown of the proteinaceous cytoskeleton by caspases.
- The cytoplasm appears dense, and the organelles appear tightly packed.
- Chromatin undergoes condensation into compact patches against the nuclear envelope in a process known as pyknosis, a hallmark of apoptosis.[24][25]
- The nuclear envelope becomes discontinuous and the DNA inside it is fragmented in a process referred to as karyorrhexis. The nucleus breaks into several discrete chromatin bodies or nucleosomal units due to the degradation of DNA.[26]
- The cell membrane shows irregular buds known as blebs.
- The cell breaks apart into several vesicles called apoptotic bodies, which are then phagocytosed.
Apoptosis progresses quickly and its products are quickly removed, making it difficult to detect or visualize. During karyorrhexis, endonuclease activation leaves short DNA fragments, regularly spaced in size. These give a characteristic "laddered" appearance on agar gel after electrophoresis. Tests for DNA laddering differentiate apoptosis from ischemic or toxic cell death.[27]
[edit] Removal of dead cells
The removal of dead cells by neighboring phagocytic cells has been termed efferocytosis.[28] Dying cells that undergo the final stages of apoptosis display phagocytotic molecules, such as phosphatidylserine, on their cell surface.[29] Phosphatidylserine is normally found on the cytosolic surface of the plasma membrane, but is redistributed during apoptosis to the extracellular surface by a hypothetical protein known as scramblase.[30] These molecules mark the cell for phagocytosis by cells possessing the appropriate receptors, such as macrophages.[31] Upon recognition, the phagocyte reorganizes its cytoskeleton for engulfment of the cell. The removal of dying cells by phagocytes occurs in an orderly manner without eliciting an inflammatory response.
[edit] Implication in disease

[edit] Defective apoptotic pathways
The many different types of apoptotic pathways contain a multitude of different biochemical components, many of them not yet understood.[32] As a pathway is more or less sequential in nature, it is a victim of causality; removing or modifying one component leads to an effect in another. In a living organism this can have disastrous effects, often in the form of disease or disorder. A discussion of every disease caused by modification of the various apoptotic pathways would be impractical, but the concept overlying each one is the same: the normal functioning of the pathway has been disrupted in such a way as to impair the ability of the cell to undergo normal apoptosis. This results in a cell that lives past its "use-by-date" and is able to replicate and pass on any faulty machinery to its progeny, increasing the likelihood of the cell becoming cancerous or diseased.
A recently-described example of this concept in action can be seen in the development of a lung cancer called NCI-H460.[33] The X-linked inhibitor of apoptosis protein (XIAP) is overexpressed in cells of the H460 cell line. XIAPs bind to the processed form of caspase-9, and suppress the activity of apoptotic activator cytochrome c, therefore overexpression leads to a decrease in the amount of pro-apoptotic agonists. As a consequence, the balance of anti-apoptotic and pro-apoptotic effectors is upset in favour of the former, and the damaged cells continue to replicate despite being directed to die.
Dysregulation of p53
The tumor-suppressor protein p53 accumulates when DNA is damaged due to a chain of biochemical factors. Part of this pathway includes alpha-interferon and beta-interferon, which induce transcription of the p53 gene and result in the increase of p53 protein level and enhancement of cancer cell-apoptosis.[34] p53 prevents the cell from replicating by stopping the cell cycle at G1, or interphase, to give the cell time to repair, however it will induce apoptosis if damage is extensive and repair efforts fail. Any disruption to the regulation of the p53 or interferon genes will result in impaired apoptosis and the possible formation of tumors.
[edit] HIV progression
The progression of the human immunodeficiency virus infection to AIDS is primarily due to the depletion of CD4+ T-helper lymphocytes, which leads to a compromised immune system. One of the mechanisms by which T-helper cells are depleted is apoptosis, which results from a series of biochemical pathways:[35]
- HIV enzymes deactivate anti-apoptotic Bcl-2 This does not directly cause cell death, but primes the cell for apoptosis should the appropriate signal be received. In parallel, these enzymes activate pro-apoptotic procaspase-8, which does directly activate the mitochondrial events of apoptosis.
- HIV may increase the level of cellular proteins which prompt Fas-mediated apoptosis.
- HIV proteins decrease the amount of CD4 glycoprotein marker present on the cell membrane.
- Released viral particles and proteins present in extracellular fluid are able to induce apoptosis in nearby "bystander" T helper cells.
- HIV decreases the production of molecules involved in marking the cell for apoptosis, giving the virus time to replicate and continue releasing apoptotic agents and virions into the surrounding tissue.
- The infected CD4+ cell may also receive the death signal from a cytotoxic T cell.
Cells may also die as a direct consequence of viral infection. HIV-1 expression induces tubular cell G2/M arrest and apoptosis. Vashistha H, Husain M, Kumar D, Yadav A, Arora S, Singhal PC. (2008)Ren Fail. 2008;30(6):655-64.
[edit] Viral infection
Viruses can trigger apoptosis of infected cells via a range of mechanisms including:
- Receptor binding.
- Activation of protein kinase R (PKR).
- Interaction with p53.
- Expression of viral proteins coupled to MHC proteins on the surface of the infected cell, allowing recognition by cells of the immune system (such as Natural Killer and cytotoxic T cells) that then induce the infected cell to undergo apoptosis.[36]
Most viruses encode proteins that can inhibit apoptosis.[37] Several viruses encode viral homologs of Bcl-2. These homologs can inhibit pro-apoptotic proteins such as BAX and BAK, which are essential for the activation of apoptosis. Examples of viral Bcl-2 proteins include the Epstein-Barr virus BHRF1 protein and the adenovirus E1B 19K protein.[38] Some viruses express caspase inhibitors that inhibit caspase activity and an example is the CrmA protein of cowpox viruses. Whilst a number of viruses can block the effects of TNF and Fas. For example the M-T2 protein of myxoma viruses can bind TNF preventing it from binding the TNF receptor and inducing a response.[39] Furthermore, many viruses express p53 inhibitors that can bind p53 and inhibit its transcriptional transactivation activity. Consequently p53 cannot induce apoptosis since it cannot induce the expression of pro-apoptotic proteins. The adenovirus E1B-55K protein and the hepatitis B virus HBx protein are examples of viral proteins that can perform such a function.[40]
Interestingly, viruses can remain intact from apoptosis particularly in the latter stages of infection. They can be exported in the apoptotic bodies that pinch off from the surface of the dying cell and the fact that they are engulfed by phagocytes prevents the initiation of a host response. This favours the spread of the virus.[39]
[edit] Apoptosis in plants
Programmed cell death in plants has a number of molecular similarities to animal apoptosis, but it also has differences, notably the presence of a cell wall and the lack of an immune system which removes the pieces of the dead cell. Instead of an immune response, the dying cell synthesizes substances to break itself down and places them in a vacuole which ruptures as the cell dies. Whether this whole process resembles animal apoptosis closely enough to warrant using the name apoptosis (as opposed to the more general programmed cell death) is unclear.[41]
[edit] Caspase Independent Apoptosis
There is an extrinsic pathway that has been noticed in several toxicity studies. It was shown that an increase in calcium concentration within a cell, caused by drug activity, also has the ability to cause apoptosis via a calcium-binding calpain protease.
[edit] Apoptosis Protein Subcellular Location Prediction
In 2003, a method was developed for predicting subcellular location of apoptosis proteins [42] Subsequently, various different modes of Chou's pseudo amino acid composition were developed for improving the quality of predcting subcellular localization of apoptosis proteins based on their sequence information alone. [43] [44] [45] [46]
[edit] See also
- Anoikis
- Apaf-1
- Apo2.7
- Apoptosis DNA Fragmentation
- Autolysis
- Autophagy
- Autophagy network
- Cisplatin
- Entosis
- Immunology
- Necrosis
- p53
- cytotoxicity
[edit] References
- ^ a b c us dict: ä�p'–É�–tÅ�'–sä�s (American Heritage Dictionary)
- ^ "About Apoptosis". http://www.nih.gov/sigs/aig/Aboutapo.html. Retrieved November 2009. "Apoptosis Interest Group, preferred pronunciation of National Institute of Health"
- ^ Kerr, JF. (1965). "A histochemical study of hypertrophy and ischaemic injury of rat liver with special reference to changes in lysosomes". Journal of Pathology and Bacteriology 90 (90): 419'35. doi:10.1002/path.1700900210.
- ^ Agency for Science, Technology and Research. "Prof Andrew H. Wyllie - Lecture Abstract". http://www.a-star.edu.sg/astar/biomed/action/biomed_dvp_abstract.do?id=2901ddeb02dH. Retrieved 2007-03-30.
- ^ Kerr JF, Wyllie AH, Currie AR (August 1972). "Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics". Br. J. Cancer 26 (4): 239'57. PMID 4561027.
- ^ O'Rourke MG, Ellem KA (2000). "John Kerr and apoptosis". Med. J. Aust. 173 (11-12): 616'7. PMID 11379508. http://www.mja.com.au/public/issues/173_11_041200/orourke/orourke.html.
- ^ Apoptosis Interest Group (1999). "About apoptosis". http://www.nih.gov/sigs/aig/Aboutapo.html. Retrieved 2006-12-15.
- ^ Webster.com dictionary entry
- ^ Popov SG, Villasmil R, Bernardi J (April 2002). "Lethal toxin of Bacillus anthracis causes apoptosis of macrophages". Biochem. Biophys. Res. Commun. 293 (1): 349'55. doi:10.1016/S0006-291X(02)00227-9. PMID 12054607.
- ^ a b Brüne B (August 2003). "Nitric oxide: NO apoptosis or turning it ON?". Cell Death Differ. 10 (8): 864'9. doi:10.1038/sj.cdd.4401261. PMID 12867993.
- ^ a b c d e f g h Cotran; Kumar, Collins (1998). Robbins Pathologic Basis of Disease. Philadelphia: W.B Saunders Company. ISBN 0-7216-7335-X.
- ^ Calcium orchestrates apoptosis. Mark P. Mattson & Sic L. Chan. Nature Cell Biology 5, 1041 - 1043 (2003). doi:10.1038/ncb1203-1041
- ^ Chiarugi A, Moskowitz MA (2002). "PARP-1'a perpetrator of apoptotic cell death?". Science 297 (5579): 259'63. doi:10.1126/science.1074592. PMID 12114611.
- ^ Fesik SW, Shi Y. (2001). "Controlling the caspases". Science 294 (5546): 1477'8. doi:10.1126/science.1062236. PMID 11711663.
- ^ Laurent M. Dejean, Sonia Martinez-Caballero, Kathleen W. Kinnally (2006). "Is MAC the knife that cuts cytochrome c from mitochondria during apoptosis?". Cell Death and Differentiation 13 (8): 1387'5. doi:10.1038/sj.cdd.4401949. PMID 16676005. http://www.nature.com/cdd/journal/v13/n8/full/4401949a.html.
- ^ Dejean LM, Martinez-Caballero S, Manon S, Kinnally KW (February 2006). "Regulation of the mitochondrial apoptosis-induced channel, MAC, by BCL-2 family proteins". Biochim. Biophys. Acta 1762 (2): 191'201. doi:10.1016/j.bbadis.2005.07.002. PMID 16055309.
- ^ Lodish, Harvey; et al. (2004). Molecular Cell Biology. New York: W.H. Freedman and Company. ISBN 0-7167-4366-3.
- ^ a b Wajant H (2002). "The Fas signaling pathway: more than a paradigm". Science 296 (5573): 1635'6. doi:10.1126/science.1071553. PMID 12040174.
- ^ Chen G, Goeddel DV (2002). "TNF-R1 signaling: a beautiful pathway". Science 296 (5573): 1634'5. doi:10.1126/science.1071924. PMID 12040173.
- ^ Goeddel, DV. "Connection Map for Tumor Necrosis Factor Pathway". Science. doi:10.1126/stke.3822007tw132] (inactive 2009-11-14). http://stke.sciencemag.org/cgi/cm/CMP_7107.
- ^ Wajant, H.. "Connection Map for Fas Signaling Pathway". Science. doi:10.1126/stke.3802007tr1] (inactive 2009-11-14). http://stke.sciencemag.org/cgi/cm/CMP_7966.
- ^ Murphy KM, Ranganathan V, Farnsworth ML, Kavallaris M, Lock RB (January 2000). "Bcl-2 inhibits Bax translocation from cytosol to mitochondria during drug-induced apoptosis of human tumor cells". Cell Death Differ. 7 (1): 102'11. doi:10.1038/sj.cdd.4400597. PMID 10713725.
- ^ Susin SA, Lorenzo HK, Zamzami N (February 1999). "Molecular characterization of mitochondrial apoptosis-inducing factor". Nature 397 (6718): 441'6. doi:10.1038/17135. PMID 9989411.
- ^ Santos A. Susin; Daugas, E; Ravagnan, L; Samejima, K; Zamzami, N; Loeffler, M; Costantini, P; Ferri, KF et al. (2000). "Two Distinct Pathways Leading to Nuclear Apoptosis". Journal of Experimental Medicine 192 (4): 571'80. doi:10.1073/pnas.191208598v1 (inactive 2009-11-14). PMID 10952727. PMC 2193229. http://www.jem.org/cgi/content/abstract/192/4/571.
- ^ Madeleine Kihlmark; Imreh, G; Hallberg, E (15 October 2001). "Sequential degradation of proteins from the nuclear envelope during apoptosis". Journal of Cell Science 114 (20): 3643'53. PMID 11707516. http://jcs.biologists.org/cgi/content/full/114/20/3643.
- ^ Nagata S (April 2000). "Apoptotic DNA fragmentation". Exp. Cell Res. 256 (1): 12'8. doi:10.1006/excr.2000.4834. PMID 10739646.
- ^ M Iwata, D Myerson, B Torok-Storb and RA Zager (1996). "An evaluation of renal tubular DNA laddering in response to oxygen deprivation and oxidant injury". http://jasn.asnjournals.org/cgi/content/abstract/5/6/1307. Retrieved 2006-04-17.
- ^ Vandivier RW, Henson PM, Douglas IS (June 2006). "Burying the dead: the impact of failed apoptotic cell removal (efferocytosis) on chronic inflammatory lung disease". Chest 129 (6): 1673'82. doi:10.1378/chest.129.6.1673. PMID 16778289.
- ^ Li MO; Sarkisian, MR; Mehal, WZ; Rakic, P; Flavell, RA (2003). "Phosphatidylserine receptor is required for clearance of apoptotic cells". Science 302 (5650): 1560'3. doi:10.1126/science.1087621. PMID 14645847.
- ^ Wang X; Wu, YC; Fadok, VA; Lee, MC; Gengyo-Ando, K; Cheng, LC; Ledwich, D; Hsu, PK et al. (2003). "Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12". Science 302 (5650): 1563'1566. doi:10.1126/science.1087641. PMID 14645848.
- ^ Savill J, Gregory C, Haslett C. (2003). "Eat me or die". Science 302 (5650): 1516'7. doi:10.1126/science.1092533. PMID 14645835.
- ^ Thompson, CB (1995). "Apoptosis in the pathogenesis and treatment of disease". Science 267 (5203): 1456'62. doi:10.1126/science.7878464. PMID 7878464.
- ^ Yang L, Mashima T, Sato S (February 2003). "Predominant suppression of apoptosome by inhibitor of apoptosis protein in non-small cell lung cancer H460 cells: therapeutic effect of a novel polyarginine-conjugated Smac peptide". Cancer Res. 63 (4): 831'7. PMID 12591734. http://cancerres.aacrjournals.org/cgi/pmidlookup?view=long&pmid=12591734.
- ^ Takaoka A; Hayakawa, S; Yanai, H; Stoiber, D; Negishi, H; Kikuchi, H; Sasaki, S; Imai, K et al. (2003). "Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence". Nature 424 (6948): 516'23. doi:10.1038/nature01850. PMID 12872134.
- ^ Judie B. Alimonti, T. Blake Ball, Keith R. Fowke (2003). "Mechanisms of CD4+ T lymphocyte cell death in human immunodeficiency virus infection and AIDS". J Gen Virology 84 (84): 1649'61. doi:10.1099/vir.0.19110-0. PMID 12810858. http://vir.sgmjournals.org/cgi/content/full/84/7/1649.
- ^ Everett, H. and McFadden, G. (1999). "Apoptosis: an innate immune response to virus infection". Trends Microbiol 7 (4): 160'5. doi:10.1016/S0966-842X(99)01487-0. PMID 10217831.
- ^ Teodoro, J.G. Branton, P.E. (1997). "Regulation of apoptosis by viral gene products". J Virol 71 (3): 1739'46. PMID 9032302.
- ^ Polster, B.M. Pevsner, J. and Hardwick, J.M. (2004). "Viral Bcl-2 homologs and their role in virus replication and associated diseases". Biochim Biophys Acta 1644 (2'3): 211'27. doi:10.1016/j.bbamcr.2003.11.001. PMID 14996505.
- ^ a b Hay, S. and Kannourakis, G. (2002). "A time to kill: viral manipulation of the cell death program". J Gen Virol 83 (Pt 7): 1547'64. PMID 12075073.
- ^ Wang, X.W. Gibson, M.K. Vermeulen, W. Yeh, H. Forrester, K. Sturzbecher, H.W. Hoeijmakers, J.H. and Harris, C.C. (1995). "Abrogation of p53-induced Apoptosis by the Hepatitis B Virus X Gene". Cancer Res 55 (24): 6012'6. PMID 8521383.
- ^ Cyrelys Collazo, Osmani Chacón, Orlando Borr�¡s (2006). "Programmed cell death in plants resembles apoptosis of animals". Biotecnología Aplicada 23: 1'10. http://elfosscientiae.cigb.edu.cu/PDFs/BA/2006/23/1/BA002301RV001-010.pdf.
- ^ Zhou, G. P. & Doctor, K. (2003). Subcellular location prediction of apoptosis proteins. PROTEINS: Structure, Function, and Genetics 50, 44-48.
- ^ Chen, Y. L. & Li, Q. Z. (2007). Prediction of apoptosis protein subcellular location using improved hybrid approach and pseudo amino acid composition. Journal of Theoretical Biology 248, 377'381.
- ^ Ding, Y. S. & Zhang, T. L. (2008). Using Chou's pseudo amino acid composition to predict subcellular localization of apoptosis proteins: an approach with immune genetic algorithm-based ensemble classifier. Pattern Recognition Letters 29, 1887-1892.
- ^ Jiang, X., Wei, R., Zhang, T. L. & Gu, Q. (2008). Using the concept of Chou's pseudo amino acid composition to predict apoptosis proteins subcellular location: an approach by approximate entropy. Protein & Peptide Letters 15, 392-396.
- ^ Lin, H., Wang, H., Ding, H., Chen, Y. L. & Li, Q. Z. (2009). Prediction of Subcellular Localization of Apoptosis Protein Using Chou's Pseudo Amino Acid Composition. Acta Biotheoretica 57, 321-330.
Vashistha H, Husain M, Kumar D, Yadav A, Arora S, Singhal PC. (2008) HIV-1 expression induces tubular cell G2/M arrest and apoptosis. Ren Fail. 2008;30(6):655-64.
[edit] External links
- Apoptosis & cell surface
- Apoptosis & Caspase 3, The Proteolysis Map-animation
- Apoptosis & Caspase 8, The Proteolysis Map-animation
- Apoptosis & Caspase 7, The Proteolysis Map-animation
- Apoptosis MiniCOPE Dictionary- list of apoptosis terms and acronyms
- Apoptosis (Programmed Cell Death) - The Virtual Library of Biochemistry and Cell Biology
- Apoptosis Research Portal
- Apoptosis Info Apoptosis protocols, articles, news, and recent publications.
- Apoptosis Signaling Pathways
- Database of proteins involved in apoptosis
- Apoptosis Video
- The Mechanisms of Apoptosis Kimball's Biology Pages. Simple explanation of the mechanisms of apoptosis triggered by internal signals (bcl-2), along the caspase-9, caspase-3 and caspase-7 pathway; and by external signals (FAS and TNF), along the caspase 8 pathway. Accessed 25 March 2007.
- WikiPathways - Apoptosis pathway
- Finding Cancer's Self-Destruct Button CR magazine (Spring 2007). Article on apoptosis and cancer.
- Apoptosis on-line lecture by Wang Xiaodong
- The Bcl-2 Family Database
- DeathBase: a database of proteins involved in cell death, curated by experts
|
||||||||||||||||
|
|||||||||||||||||||||||||
|
||||||||||||||||||||||||||
|
SOURCES.COM is an online portal and directory for journalists, news media, researchers and anyone seeking experts, spokespersons, and reliable information resources. Use SOURCES.COM to find experts, media contacts, news releases, background information, scientists, officials, speakers, newsmakers, spokespeople, talk show guests, story ideas, research studies, databases, universities, associations and NGOs, businesses, government spokespeople. Indexing and search applications by Ulli Diemer and Chris DeFreitas.
For information about being included in SOURCES as a expert or spokesperson see the FAQ . For partnerships, content and applications, and domain name opportunities contact us.